DNA polymerase beta
| POLB | |||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Identifiers | |||||||||||||||||||||||||||||||||||||||||||||||||||
| Aliases | POLB, DNA polymerase beta | ||||||||||||||||||||||||||||||||||||||||||||||||||
| External IDs | OMIM: 174760; MGI: 97740; HomoloGene: 2013; GeneCards: POLB; OMA:POLB - orthologs | ||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Wikidata | |||||||||||||||||||||||||||||||||||||||||||||||||||
| |||||||||||||||||||||||||||||||||||||||||||||||||||
| Stem loopII regulatory element in POLB | |
|---|---|
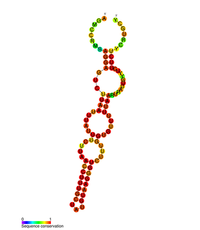 Predicted secondary structure of the stem loopII (M2) regulatory element in POLB | |
| Identifiers | |
| Symbol | POLB |
| Rfam | RF01455 |
| NCBI Gene | 5423 |
| HGNC | 9174 |
| OMIM | 174760 |
| RefSeq | NM_002690 |
| Other data | |
| RNA type | Cis-reg |
| Domain(s) | Mammalia |
| Locus | Chr. 8 p11.2 |
| PDB structures | PDBe |
DNA polymerase beta, also known as POLB, is an enzyme present in eukaryotes. In humans, it is encoded by the POLB gene.[4]
Function
In eukaryotic cells, DNA polymerase beta (POLB) performs base excision repair (BER) required for DNA maintenance, replication, recombination, and drug resistance.[4]
The mitochondrial DNA of mammalian cells is constantly under attack from oxygen radicals released during ATP production. Mammalian cell mitochondria contain an efficient base excision repair system employing POLB that removes some frequent oxidative DNA damages.[5] POLB thus has a key role in maintaining the stability of the mitochondrial genome.[5]
An analysis of the fidelity of DNA replication by polymerase beta in the neurons from young and very aged mice indicated that aging has no significant effect on the fidelity of DNA synthesis by polymerase beta.[6] This finding was considered to provide evidence against the error catastrophe theory of aging.[6][7]
Base excision repair
Cabelof et al. measured the ability to repair DNA damage by the BER pathway in tissues of young (4-month-old) and old (24-month-old) mice.[8] In all tissues examined (brain, liver, spleen and testes) the ability to repair DNA damage declined significantly with age, and the reduction in repair capability correlated with decreased levels of DNA polymerase beta at both the protein and messenger RNA levels. Numerous investigators have reported an accumulation of DNA damage with age, especially in brain and liver.[9] Cabelof et al.[8] suggested that the inability of the BER pathway to repair damages over time may provide a mechanistic explanation for the frequent observations of DNA accumulation of damage with age.
Regulation of expression
DNA polymerase beta maintains genome integrity by participating in base excision repair. Overexpression of POLB mRNA has been correlated with a number of cancer types, whereas deficiencies in POLB results in hypersensitivity to alkylating agents, induced apoptosis, and chromosomal breaking. Therefore, it is essential that POLB expression is tightly regulated.[10][11][12][13]
POLB gene is upregulated by CREB1 transcription factor's binding to the cAMP response element (CRE) present in the promoter of the POLB gene in response to exposure to alkylating agents.[14][15] POLB gene expression is also regulated at the post transcriptional level as the 3'UTR of the POLB mRNA has been shown to contain three stem-loop structures that influence gene expression.[16] These three-stem loop structures are known as M1, M2, and M3, where M2 and M3 have a key role in gene regulation. M3 contributes to gene expression, as it contains the polyadenylation signal followed by the cleavage and polyadenylation site, thereby contributing to pre-mRNA processing. M2 has been shown to be evolutionary conserved, and, through mutagenesis, it was shown that this stem loop structure acts as a RNA destabilizing element.
In addition to these cis-regulatory elements present within the 3'UTR a trans-acting protein, HAX1 is thought to contribute to the regulation of gene expression. Yeast three-hybrid assays have shown that this protein binds to the stem loops within the 3'UTR of the POLB mRNA, however the exact mechanism in how this protein regulates gene expression is still to be determined.
Interactions
DNA polymerase beta has been shown to interact with PNKP[17] and XRCC1.[18][19][20][21]
See also
- POLA1
- POLA2
References
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000031536 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ a b "NCBI Gene: DNA polymerase beta".
- ^ a b Prasad R, Çağlayan M, Dai DP, Nadalutti CA, Zhao ML, Gassman NR, et al. (December 2017). "DNA polymerase β: A missing link of the base excision repair machinery in mammalian mitochondria". DNA Repair. 60: 77–88. doi:10.1016/j.dnarep.2017.10.011. PMC 5919216. PMID 29100041.
- ^ a b Subba Rao K, Martin GM, Loeb LA (October 1985). "Fidelity of DNA polymerase-beta in neurons from young and very aged mice". Journal of Neurochemistry. 45 (4): 1273–8. doi:10.1111/j.1471-4159.1985.tb05553.x. PMID 3161998. S2CID 84448241.
- ^ Orgel LE (June 1973). "Ageing of clones of mammalian cells". Nature. 243 (5408): 441–5. Bibcode:1973Natur.243..441O. doi:10.1038/243441a0. PMID 4591306. S2CID 4153800.
- ^ a b Cabelof DC, Raffoul JJ, Yanamadala S, Ganir C, Guo Z, Heydari AR (March 2002). "Attenuation of DNA polymerase beta-dependent base excision repair and increased DMS-induced mutagenicity in aged mice". Mutation Research. 500 (1–2): 135–45. doi:10.1016/s0027-5107(02)00003-9. PMC 3339152. PMID 11890943.
- ^ Bernstein C, Bernstein H (1991). Aging, Sex, and DNA Repair. San Diego: Academic Press. pp. 46–60. ISBN 0-12-092860-4.
- ^ Canitrot Y, Cazaux C, Fréchet M, Bouayadi K, Lesca C, Salles B, Hoffmann JS (October 1998). "Overexpression of DNA polymerase beta in cell results in a mutator phenotype and a decreased sensitivity to anticancer drugs". Proceedings of the National Academy of Sciences of the United States of America. 95 (21): 12586–90. Bibcode:1998PNAS...9512586C. doi:10.1073/pnas.95.21.12586. PMC 22874. PMID 9770529.
- ^ Bergoglio V, Pillaire MJ, Lacroix-Triki M, Raynaud-Messina B, Canitrot Y, Bieth A, et al. (June 2002). "Deregulated DNA polymerase beta induces chromosome instability and tumorigenesis". Cancer Research. 62 (12): 3511–4. PMID 12067997.
- ^ Bergoglio V, Canitrot Y, Hogarth L, Minto L, Howell SB, Cazaux C, Hoffmann JS (September 2001). "Enhanced expression and activity of DNA polymerase beta in human ovarian tumor cells: impact on sensitivity towards antitumor agents". Oncogene. 20 (43): 6181–7. doi:10.1038/sj.onc.1204743. PMID 11593426. S2CID 30039904.
- ^ Srivastava DK, Husain I, Arteaga CL, Wilson SH (June 1999). "DNA polymerase beta expression differences in selected human tumors and cell lines". Carcinogenesis. 20 (6): 1049–54. doi:10.1093/carcin/20.6.1049. PMID 10357787.
- ^ He F, Yang XP, Srivastava DK, Wilson SH (January 2003). "DNA polymerase beta gene expression: the promoter activator CREB-1 is upregulated in Chinese hamster ovary cells by DNA alkylating agent-induced stress". Biological Chemistry. 384 (1): 19–23. doi:10.1515/BC.2003.003. PMID 12674496. S2CID 33798724.
- ^ Narayan S, He F, Wilson SH (August 1996). "Activation of the human DNA polymerase beta promoter by a DNA-alkylating agent through induced phosphorylation of cAMP response element-binding protein-1". The Journal of Biological Chemistry. 271 (31): 18508–13. doi:10.1074/jbc.271.31.18508. PMID 8702497.
- ^ Sarnowska E, Grzybowska EA, Sobczak K, Konopinski R, Wilczynska A, Szwarc M, et al. (2007). "Hairpin structure within the 3'UTR of DNA polymerase beta mRNA acts as a post-transcriptional regulatory element and interacts with Hax-1". Nucleic Acids Research. 35 (16): 5499–510. doi:10.1093/nar/gkm502. PMC 2018635. PMID 17704138.
- ^ Whitehouse CJ, Taylor RM, Thistlethwaite A, Zhang H, Karimi-Busheri F, Lasko DD, et al. (January 2001). "XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair". Cell. 104 (1): 107–17. doi:10.1016/S0092-8674(01)00195-7. PMID 11163244. S2CID 1487128.
- ^ Wang L, Bhattacharyya N, Chelsea DM, Escobar PF, Banerjee S (November 2004). "A novel nuclear protein, MGC5306 interacts with DNA polymerase beta and has a potential role in cellular phenotype". Cancer Research. 64 (21): 7673–7. doi:10.1158/0008-5472.CAN-04-2801. PMID 15520167. S2CID 35569215.
- ^ Fan J, Otterlei M, Wong HK, Tomkinson AE, Wilson DM (2004). "XRCC1 co-localizes and physically interacts with PCNA". Nucleic Acids Research. 32 (7): 2193–201. doi:10.1093/nar/gkh556. PMC 407833. PMID 15107487.
- ^ Kubota Y, Nash RA, Klungland A, Schär P, Barnes DE, Lindahl T (December 1996). "Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein". The EMBO Journal. 15 (23): 6662–70. doi:10.1002/j.1460-2075.1996.tb01056.x. PMC 452490. PMID 8978692.
- ^ Bhattacharyya N, Banerjee S (July 2001). "A novel role of XRCC1 in the functions of a DNA polymerase beta variant". Biochemistry. 40 (30): 9005–13. doi:10.1021/bi0028789. PMID 11467963.
Further reading
- Date T, Tanihara K, Yamamoto S, Nomura N, Matsukage A (September 1992). "Two regions in human DNA polymerase beta mRNA suppress translation in Escherichia coli". Nucleic Acids Research. 20 (18): 4859–64. doi:10.1093/nar/20.18.4859. PMC 334243. PMID 1408801.
- Wang L, Patel U, Ghosh L, Banerjee S (September 1992). "DNA polymerase beta mutations in human colorectal cancer". Cancer Research. 52 (17): 4824–7. PMID 1511447.
- Tokui T, Inagaki M, Nishizawa K, Yatani R, Kusagawa M, Ajiro K, et al. (June 1991). "Inactivation of DNA polymerase beta by in vitro phosphorylation with protein kinase C". The Journal of Biological Chemistry. 266 (17): 10820–4. doi:10.1016/S0021-9258(18)99092-7. PMID 2040602.
- SenGupta DN, Zmudzka BZ, Kumar P, Cobianchi F, Skowronski J, Wilson SH (April 1986). "Sequence of human DNA polymerase beta mRNA obtained through cDNA cloning". Biochemical and Biophysical Research Communications. 136 (1): 341–7. doi:10.1016/0006-291X(86)90916-2. PMID 2423078.
- Zmudzka BZ, Fornace A, Collins J, Wilson SH (October 1988). "Characterization of DNA polymerase beta mRNA: cell-cycle and growth response in cultured human cells". Nucleic Acids Research. 16 (20): 9587–96. doi:10.1093/nar/16.20.9587. PMC 338765. PMID 2460824.
- Widen SG, Kedar P, Wilson SH (November 1988). "Human beta-polymerase gene. Structure of the 5'-flanking region and active promoter". The Journal of Biological Chemistry. 263 (32): 16992–8. doi:10.1016/S0021-9258(18)37488-X. PMID 3182828.
- Abbotts J, SenGupta DN, Zmudzka B, Widen SG, Notario V, Wilson SH (February 1988). "Expression of human DNA polymerase beta in Escherichia coli and characterization of the recombinant enzyme". Biochemistry. 27 (3): 901–9. doi:10.1021/bi00403a010. PMID 3284575.
- Dobashi Y, Kubota Y, Shuin T, Torigoe S, Yao M, Hosaka M (April 1995). "Polymorphisms in the human DNA polymerase beta gene". Human Genetics. 95 (4): 389–90. doi:10.1007/bf00208961. PMID 7705833. S2CID 22658070.
- Chyan YJ, Ackerman S, Shepherd NS, McBride OW, Widen SG, Wilson SH, Wood TG (July 1994). "The human DNA polymerase beta gene structure. Evidence of alternative splicing in gene expression". Nucleic Acids Research. 22 (14): 2719–25. doi:10.1093/nar/22.14.2719. PMC 308239. PMID 7914364.
- Maruyama K, Sugano S (January 1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Chang M, Burmer GC, Sweasy J, Loeb LA, Edelhoff S, Disteche CM, et al. (May 1994). "Evidence against DNA polymerase beta as a candidate gene for Werner syndrome". Human Genetics. 93 (5): 507–12. doi:10.1007/bf00202813. PMID 8168825. S2CID 34824853.
- Chyan YJ, Strauss PR, Wood TG, Wilson SH (August 1996). "Identification of novel mRNA isoforms for human DNA polymerase beta". DNA and Cell Biology. 15 (8): 653–9. doi:10.1089/dna.1996.15.653. PMID 8769567.
- Pelletier H, Sawaya MR, Wolfle W, Wilson SH, Kraut J (October 1996). "Crystal structures of human DNA polymerase beta complexed with DNA: implications for catalytic mechanism, processivity, and fidelity". Biochemistry. 35 (39): 12742–61. doi:10.1021/bi952955d. PMID 8841118.
- Pelletier H, Sawaya MR, Wolfle W, Wilson SH, Kraut J (October 1996). "A structural basis for metal ion mutagenicity and nucleotide selectivity in human DNA polymerase beta". Biochemistry. 35 (39): 12762–77. doi:10.1021/bi9529566. PMID 8841119.
- Pelletier H, Sawaya MR (October 1996). "Characterization of the metal ion binding helix-hairpin-helix motifs in human DNA polymerase beta by X-ray structural analysis". Biochemistry. 35 (39): 12778–87. doi:10.1021/bi960790i. PMID 8841120.
- Kubota Y, Nash RA, Klungland A, Schär P, Barnes DE, Lindahl T (December 1996). "Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein". The EMBO Journal. 15 (23): 6662–70. doi:10.1002/j.1460-2075.1996.tb01056.x. PMC 452490. PMID 8978692.
- Bennett RA, Wilson DM, Wong D, Demple B (July 1997). "Interaction of human apurinic endonuclease and DNA polymerase beta in the base excision repair pathway". Proceedings of the National Academy of Sciences of the United States of America. 94 (14): 7166–9. Bibcode:1997PNAS...94.7166B. doi:10.1073/pnas.94.14.7166. PMC 23779. PMID 9207062.
- Sawaya MR, Prasad R, Wilson SH, Kraut J, Pelletier H (September 1997). "Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism". Biochemistry. 36 (37): 11205–15. doi:10.1021/bi9703812. PMID 9287163.
- Bhattacharyya N, Banerjee S (September 1997). "A variant of DNA polymerase beta acts as a dominant negative mutant". Proceedings of the National Academy of Sciences of the United States of America. 94 (19): 10324–9. Bibcode:1997PNAS...9410324B. doi:10.1073/pnas.94.19.10324. PMC 23361. PMID 9294209.
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S (October 1997). "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library". Gene. 200 (1–2): 149–56. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
- Tan XH, Zhao M, Pan KF, Dong Y, Dong B, Feng GJ, et al. (March 2005). "Frequent mutation related with overexpression of DNA polymerase beta in primary tumors and precancerous lesions of human stomach". Cancer Letters. 220 (1): 101–14. doi:10.1016/j.canlet.2004.07.049. PMID 15737693.
External links
- Rfam entry for the stem loopII (M2) regulatory element in POLB
- v
- t
- e
-
 1bno: NMR SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA, MINIMIZED AVERAGE STRUCTURE
1bno: NMR SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA, MINIMIZED AVERAGE STRUCTURE -
 1bnp: NMR SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA, 55 STRUCTURES
1bnp: NMR SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA, 55 STRUCTURES -
 1bpb: CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA: EVIDENCE FOR A COMMON POLYMERASE MECHANISM
1bpb: CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA: EVIDENCE FOR A COMMON POLYMERASE MECHANISM -
 1bpd: CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA: EVIDENCE FOR A COMMON POLYMERASE MECHANISM
1bpd: CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA: EVIDENCE FOR A COMMON POLYMERASE MECHANISM -
 1bpe: CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA; EVIDENCE FOR A COMMON POLYMERASE MECHANISM
1bpe: CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA; EVIDENCE FOR A COMMON POLYMERASE MECHANISM -
 1bpx: DNA POLYMERASE BETA/DNA COMPLEX
1bpx: DNA POLYMERASE BETA/DNA COMPLEX -
 1bpy: HUMAN DNA POLYMERASE BETA COMPLEXED WITH GAPPED DNA AND DDCTP
1bpy: HUMAN DNA POLYMERASE BETA COMPLEXED WITH GAPPED DNA AND DDCTP -
 1bpz: HUMAN DNA POLYMERASE BETA COMPLEXED WITH NICKED DNA
1bpz: HUMAN DNA POLYMERASE BETA COMPLEXED WITH NICKED DNA -
 1dk2: REFINED SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA
1dk2: REFINED SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA -
 1dk3: REFINED SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA
1dk3: REFINED SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA -
 1huo: CRYSTAL STRUCTURE OF DNA POLYMERASE BETA COMPLEXED WITH DNA AND CR-TMPPCP
1huo: CRYSTAL STRUCTURE OF DNA POLYMERASE BETA COMPLEXED WITH DNA AND CR-TMPPCP -
 1huz: CRYSTAL STRUCTURE OF DNA POLYMERASE COMPLEXED WITH DNA AND CR-PCP
1huz: CRYSTAL STRUCTURE OF DNA POLYMERASE COMPLEXED WITH DNA AND CR-PCP -
 1jn3: FIDELITY PROPERTIES AND STRUCTURE OF M282L MUTATOR MUTANT OF DNA POLYMERASE: SUBTLE STRUCTURAL CHANGES INFLUENCE THE MECHANISM OF NUCLEOTIDE DISCRIMINATION
1jn3: FIDELITY PROPERTIES AND STRUCTURE OF M282L MUTATOR MUTANT OF DNA POLYMERASE: SUBTLE STRUCTURAL CHANGES INFLUENCE THE MECHANISM OF NUCLEOTIDE DISCRIMINATION -
 1mq2: Human DNA Polymerase Beta Complexed With Gapped DNA Containing an 8-oxo-7,8-dihydro-Guanine and dAMP
1mq2: Human DNA Polymerase Beta Complexed With Gapped DNA Containing an 8-oxo-7,8-dihydro-Guanine and dAMP -
 1mq3: Human DNA Polymerase Beta Complexed With Gapped DNA Containing an 8-oxo-7,8-dihydro-Guanine Template Paired with dCTP
1mq3: Human DNA Polymerase Beta Complexed With Gapped DNA Containing an 8-oxo-7,8-dihydro-Guanine Template Paired with dCTP -
 1nom: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7), 31-KD DOMAIN; SOAKED IN THE PRESENCE OF MNCL2 (5 MILLIMOLAR)
1nom: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7), 31-KD DOMAIN; SOAKED IN THE PRESENCE OF MNCL2 (5 MILLIMOLAR) -
 1rpl: 2.3 ANGSTROMS CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF DNA POLYMERASE BETA
1rpl: 2.3 ANGSTROMS CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF DNA POLYMERASE BETA -
 1tv9: HUMAN DNA POLYMERASE BETA COMPLEXED WITH NICKED DNA CONTAINING A MISMATCHED TEMPLATE ADENINE AND INCOMING CYTIDINE
1tv9: HUMAN DNA POLYMERASE BETA COMPLEXED WITH NICKED DNA CONTAINING A MISMATCHED TEMPLATE ADENINE AND INCOMING CYTIDINE -
 1tva: HUMAN DNA POLYMERASE BETA COMPLEXED WITH NICKED DNA CONTAINING A MISMATCHED TEMPLATE THYMIDINE AND INCOMING CYTIDINE
1tva: HUMAN DNA POLYMERASE BETA COMPLEXED WITH NICKED DNA CONTAINING A MISMATCHED TEMPLATE THYMIDINE AND INCOMING CYTIDINE -
 1zjm: Human DNA Polymerase beta complexed with DNA containing an A-A mismatched primer terminus
1zjm: Human DNA Polymerase beta complexed with DNA containing an A-A mismatched primer terminus -
 1zjn: Human DNA Polymerase beta complexed with DNA containing an A-A mismatched primer terminus with dGTP
1zjn: Human DNA Polymerase beta complexed with DNA containing an A-A mismatched primer terminus with dGTP -
 1zqa: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF KCL (150 MILLIMOLAR) AT PH 7.5
1zqa: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF KCL (150 MILLIMOLAR) AT PH 7.5 -
 1zqb: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF BACL2 (150 MILLIMOLAR)
1zqb: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF BACL2 (150 MILLIMOLAR) -
 1zqc: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CACL2 (15 MILLIMOLAR)
1zqc: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CACL2 (15 MILLIMOLAR) -
 1zqd: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CACL2 (150 MILLIMOLAR)
1zqd: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CACL2 (150 MILLIMOLAR) -
 1zqe: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CRCL3 (SATURATED SOLUTION)
1zqe: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CRCL3 (SATURATED SOLUTION) -
 1zqf: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CSCL (150 MILLIMOLAR)
1zqf: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CSCL (150 MILLIMOLAR) -
 1zqg: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF A SODIUM-FREE ARTIFICIAL MOTHER LIQUOR AT PH 6.5
1zqg: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF A SODIUM-FREE ARTIFICIAL MOTHER LIQUOR AT PH 6.5 -
 1zqh: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF A SODIUM-FREE ARTIFICIAL MOTHER LIQUOR AT PH 7.5
1zqh: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF A SODIUM-FREE ARTIFICIAL MOTHER LIQUOR AT PH 7.5 -
 1zqi: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF KCL (150 MILLIMOLAR)
1zqi: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF KCL (150 MILLIMOLAR) -
 1zqj: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CACL2 (15 MILLIMOLAR) AND MGCL2 (15 MILLIMOLAR)
1zqj: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CACL2 (15 MILLIMOLAR) AND MGCL2 (15 MILLIMOLAR) -
 1zqk: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF KCL (75 MILLIMOLAR) AND MGCL2 (75 MILLIMOLAR)
1zqk: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF KCL (75 MILLIMOLAR) AND MGCL2 (75 MILLIMOLAR) -
 1zql: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF MNCL2 (15 MILLIMOLAR) AND MGCL2 (15 MILLIMOLAR)
1zql: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF MNCL2 (15 MILLIMOLAR) AND MGCL2 (15 MILLIMOLAR) -
 1zqm: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF MNCL2 (15 MILLIMOLAR)
1zqm: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF MNCL2 (15 MILLIMOLAR) -
 1zqn: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF BACL2 (15 MILLIMOLAR) AND NACL (15 MILLIMOLAR)
1zqn: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF BACL2 (15 MILLIMOLAR) AND NACL (15 MILLIMOLAR) -
 1zqo: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CACL2 (15 MILLIMOLAR) AND NACL (15 MILLIMOLAR)
1zqo: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CACL2 (15 MILLIMOLAR) AND NACL (15 MILLIMOLAR) -
 1zqp: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF KCL (75 MILLIMOLAR) AND NACL (75 MILLIMOLAR)
1zqp: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF KCL (75 MILLIMOLAR) AND NACL (75 MILLIMOLAR) -
 1zqq: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF MNCL2 (15 MILLIMOLAR) AND NACL (15 MILLIMOLAR)
1zqq: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF MNCL2 (15 MILLIMOLAR) AND NACL (15 MILLIMOLAR) -
 1zqr: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF NICL2
1zqr: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF NICL2 -
 1zqs: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF TLCL (0.5 MILLIMOLAR)
1zqs: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF TLCL (0.5 MILLIMOLAR) -
 1zqt: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (0.01 MILLIMOLAR) AND ZNCL2 (0.02 MILLIMOLAR)
1zqt: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (0.01 MILLIMOLAR) AND ZNCL2 (0.02 MILLIMOLAR) -
 1zqu: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7), 31-KD DOMAIN; SOAKED IN THE PRESENCE OF ARTIFICIAL MOTHER LIQUOR
1zqu: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7), 31-KD DOMAIN; SOAKED IN THE PRESENCE OF ARTIFICIAL MOTHER LIQUOR -
 1zqv: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7), 31-KD DOMAIN; SOAKED IN THE PRESENCE OF CACL2 (150 MILLIMOLAR)
1zqv: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7), 31-KD DOMAIN; SOAKED IN THE PRESENCE OF CACL2 (150 MILLIMOLAR) -
 1zqw: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7), 31-KD DOMAIN; SOAKED IN THE PRESENCE OF CSCL (150 MILLIMOLAR)
1zqw: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7), 31-KD DOMAIN; SOAKED IN THE PRESENCE OF CSCL (150 MILLIMOLAR) -
 1zqx: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7), 31-KD DOMAIN; SOAKED IN THE PRESENCE OF KCL (150 MILLIMOLAR)
1zqx: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7), 31-KD DOMAIN; SOAKED IN THE PRESENCE OF KCL (150 MILLIMOLAR) -
 1zqy: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7), 31-KD DOMAIN; SOAKED IN THE PRESENCE OF MGCL2 (50 MILLIMOLAR)
1zqy: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7), 31-KD DOMAIN; SOAKED IN THE PRESENCE OF MGCL2 (50 MILLIMOLAR) -
 1zqz: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7), 31-KD DOMAIN; SOAKED IN THE PRESENCE OF MNCL2 (50 MILLIMOLAR)
1zqz: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7), 31-KD DOMAIN; SOAKED IN THE PRESENCE OF MNCL2 (50 MILLIMOLAR) -
 2bpc: CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA: EVIDENCE FOR A COMMON POLYMERASE MECHANISM
2bpc: CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA: EVIDENCE FOR A COMMON POLYMERASE MECHANISM -
 2bpf: STRUCTURES OF TERNARY COMPLEXES OF RAT DNA POLYMERASE BETA, A DNA TEMPLATE-PRIMER, AND DDCTP
2bpf: STRUCTURES OF TERNARY COMPLEXES OF RAT DNA POLYMERASE BETA, A DNA TEMPLATE-PRIMER, AND DDCTP -
 2bpg: STRUCTURES OF TERNARY COMPLEXES OF RAT DNA POLYMERASE BETA, A DNA TEMPLATE-PRIMER, AND DDCTP
2bpg: STRUCTURES OF TERNARY COMPLEXES OF RAT DNA POLYMERASE BETA, A DNA TEMPLATE-PRIMER, AND DDCTP -
 2fmp: DNA Polymerase beta with a terminated gapped DNA substrate and ddCTP with sodium in the catalytic site
2fmp: DNA Polymerase beta with a terminated gapped DNA substrate and ddCTP with sodium in the catalytic site -
 2fmq: Sodium in active site of DNA Polymerase Beta
2fmq: Sodium in active site of DNA Polymerase Beta -
 2fms: DNA Polymerase beta with a gapped DNA substrate and dUMPNPP with magnesium in the catalytic site
2fms: DNA Polymerase beta with a gapped DNA substrate and dUMPNPP with magnesium in the catalytic site -
![2i9g: DNA Polymerase Beta with a Benzo[c]phenanthrene diol epoxide adducted guanine base](//upload.wikimedia.org/wikipedia/commons/thumb/4/4c/PDB_2i9g_EBI.jpg/180px-PDB_2i9g_EBI.jpg) 2i9g: DNA Polymerase Beta with a Benzo[c]phenanthrene diol epoxide adducted guanine base
2i9g: DNA Polymerase Beta with a Benzo[c]phenanthrene diol epoxide adducted guanine base -
 2iso: Ternary complex of DNA Polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-difluoromethylene triphosphate
2iso: Ternary complex of DNA Polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-difluoromethylene triphosphate -
 2isp: Ternary complex of DNA Polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-methylene triphosphate
2isp: Ternary complex of DNA Polymerase beta with a dideoxy terminated primer and 2'-deoxyguanosine 5'-beta, gamma-methylene triphosphate -
 2p66: Human DNA Polymerase beta complexed with tetrahydrofuran (abasic site) containing DNA
2p66: Human DNA Polymerase beta complexed with tetrahydrofuran (abasic site) containing DNA -
 7ice: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF CACL2
7ice: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF CACL2 -
 7icf: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CDCL2 (0.1 MILLIMOLAR) (FOUR-DAY SOAK)
7icf: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CDCL2 (0.1 MILLIMOLAR) (FOUR-DAY SOAK) -
 7icg: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF CDCL2
7icg: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF CDCL2 -
 7ich: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF COCL2
7ich: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF COCL2 -
 7ici: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CRCL3 (0.1 MILLIMOLAR)
7ici: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CRCL3 (0.1 MILLIMOLAR) -
 7icj: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CUCL2 (0.1 MILLIMOLAR)
7icj: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CUCL2 (0.1 MILLIMOLAR) -
 7ick: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF MGCL2
7ick: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF MGCL2 -
 7icl: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF MNCL2 (0.1 MILLIMOLAR)
7icl: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF MNCL2 (0.1 MILLIMOLAR) -
 7icm: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF MNCL2 (1.0 MILLIMOLAR)
7icm: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF MNCL2 (1.0 MILLIMOLAR) -
 7icn: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF NICL2
7icn: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF NICL2 -
 7ico: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF ZNCL2
7ico: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF ZNCL2 -
 7icp: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF ZNCL2 (0.01 MILLIMOLAR)
7icp: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF ZNCL2 (0.01 MILLIMOLAR) -
 7icq: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF ZNCL2
7icq: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF ZNCL2 -
 7icr: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF ZNCL2
7icr: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF ZNCL2 -
 7ics: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF ZNCL2
7ics: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF ZNCL2 -
 7ict: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF ZNCL2 AND MGCL2
7ict: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF ZNCL2 AND MGCL2 -
 7icu: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CDCL2 (0.1 MILLIMOLAR)
7icu: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF CDCL2 (0.1 MILLIMOLAR) -
 7icv: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF MNCL2 (0.1 MILLIMOLAR) AND IN THE ABSENCE OF NACL
7icv: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF MNCL2 (0.1 MILLIMOLAR) AND IN THE ABSENCE OF NACL -
 8ica: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR) AND CACL2 (5 MILLIMOLAR)
8ica: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR) AND CACL2 (5 MILLIMOLAR) -
 8icb: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF ARTIFICIAL MOTHER LIQUOR
8icb: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF ARTIFICIAL MOTHER LIQUOR -
 8icc: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA (NO 5'-PHOSPHATE)
8icc: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA (NO 5'-PHOSPHATE) -
 8ice: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR) AND CDCL2 (1 MILLIMOLAR)
8ice: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR) AND CDCL2 (1 MILLIMOLAR) -
 8icf: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (10 MILLIMOLAR) AND MGCL2 (50 MILLIMOLAR)
8icf: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (10 MILLIMOLAR) AND MGCL2 (50 MILLIMOLAR) -
 8icg: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR) AND MGCL2 (5 MILLIMOLAR)
8icg: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR) AND MGCL2 (5 MILLIMOLAR) -
 8ich: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DCTP (1 MILLIMOLAR) AND MGCL2 (5 MILLIMOLAR)
8ich: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DCTP (1 MILLIMOLAR) AND MGCL2 (5 MILLIMOLAR) -
 8ici: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DGTP (1 MILLIMOLAR) AND MGCL2 (5 MILLIMOLAR)
8ici: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DGTP (1 MILLIMOLAR) AND MGCL2 (5 MILLIMOLAR) -
 8icj: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + THYMIDINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DTTP AND MGCL2
8icj: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + THYMIDINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DTTP AND MGCL2 -
 8ick: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR), MGCL2 (5 MILLIMOLAR), AND MNCL2 (5 MILLIMOLAR)
8ick: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR), MGCL2 (5 MILLIMOLAR), AND MNCL2 (5 MILLIMOLAR) -
 8icl: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR) AND NICL2 (5 MILLIMOLAR)
8icl: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR) AND NICL2 (5 MILLIMOLAR) -
 8icm: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR), MNCL2 (5 MILLIMOLAR), AND AMMONIUM SULFATE (75 MILLIMOLAR)
8icm: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR), MNCL2 (5 MILLIMOLAR), AND AMMONIUM SULFATE (75 MILLIMOLAR) -
 8icn: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF ATP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR)
8icn: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF ATP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR) -
 8ico: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF AZT-TP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR)
8ico: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF AZT-TP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR) -
 8icp: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR)
8icp: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR) -
 8icq: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (0.1 MILLIMOLAR) AND MNCL2 (0.5 MILLIMOLAR)
8icq: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (0.1 MILLIMOLAR) AND MNCL2 (0.5 MILLIMOLAR) -
 8icr: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR)
8icr: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR) -
 8ics: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DCTP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR)
8ics: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DCTP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR) -
 8ict: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DCTP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR)
8ict: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DCTP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR) -
 8icu: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DDATP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR)
8icu: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DDATP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR) -
 8icv: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DGTP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR)
8icv: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DGTP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR) -
 8icw: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DTTP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR)
8icw: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DTTP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR) -
 8icx: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DTTP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR)
8icx: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DTTP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR) -
 8icy: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + THYMIDINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DTTP AND MNCL2
8icy: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + THYMIDINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DTTP AND MNCL2 -
 8icz: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF OF DATP (1 MILLIMOLAR), MNCL2 (5 MILLIMOLAR), AND LITHIUM SULFATE (75 MILLIMOLAR)
8icz: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF OF DATP (1 MILLIMOLAR), MNCL2 (5 MILLIMOLAR), AND LITHIUM SULFATE (75 MILLIMOLAR) -
 9ica: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2'-DEOXYADENOSINE-5'-O-(1-THIOTRIPHOSPHATE), SOAKED IN THE PRESENCE OF DATP(ALPHA)S AND MNCL2
9ica: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2'-DEOXYADENOSINE-5'-O-(1-THIOTRIPHOSPHATE), SOAKED IN THE PRESENCE OF DATP(ALPHA)S AND MNCL2 -
 9icb: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2'-DEOXYADENOSINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DATP AND COCL2
9icb: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2'-DEOXYADENOSINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DATP AND COCL2 -
 9icc: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2'-DEOXYADENOSINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DATP AND CRCL3
9icc: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2'-DEOXYADENOSINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DATP AND CRCL3 -
 9ice: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR) AND CUCL2 (0.1 MILLIMOLAR)
9ice: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR) AND CUCL2 (0.1 MILLIMOLAR) -
 9icf: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2'-DEOXYADENOSINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DATP AND ZNCL2
9icf: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2'-DEOXYADENOSINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DATP AND ZNCL2 -
 9icg: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DCTP (1 MILLIMOLAR) AND ZNCL2 (1 MILLIMOLAR)
9icg: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DCTP (1 MILLIMOLAR) AND ZNCL2 (1 MILLIMOLAR) -
 9ich: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DGTP (1 MILLIMOLAR) AND ZNCL2 (1 MILLIMOLAR)
9ich: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DGTP (1 MILLIMOLAR) AND ZNCL2 (1 MILLIMOLAR) -
 9ici: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DTTP (1 MILLIMOLAR) AND ZNCL2 (1 MILLIMOLAR)
9ici: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DTTP (1 MILLIMOLAR) AND ZNCL2 (1 MILLIMOLAR) -
 9icj: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA
9icj: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA -
 9ick: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF ARTIFICIAL MOTHER LIQUOR
9ick: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF ARTIFICIAL MOTHER LIQUOR -
 9icl: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF PYROPHOSPHATE AND MNCL2
9icl: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF PYROPHOSPHATE AND MNCL2 -
 9icm: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DOUBLE STRANDED DNA (NO 5'-PHOSPHATE)
9icm: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DOUBLE STRANDED DNA (NO 5'-PHOSPHATE) -
 9icn: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2',3'-DIDEOXYCYTIDINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DDCTP AND MGCL2
9icn: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2',3'-DIDEOXYCYTIDINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DDCTP AND MGCL2 -
 9ico: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF DTTP AND MGCL2
9ico: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX, SOAKED IN THE PRESENCE OF DTTP AND MGCL2 -
 9icp: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF PYROPHOSPHATE (1 MILLIMOLAR) AND MGCL2 (5 MILLIMOLAR)
9icp: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF PYROPHOSPHATE (1 MILLIMOLAR) AND MGCL2 (5 MILLIMOLAR) -
 9icq: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR)
9icq: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DATP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR) -
 9icr: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DCTP AND MNCL2
9icr: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DCTP AND MNCL2 -
 9ics: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2',3'-DIDEOXYCYTIDINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DDCTP AND MNCL2
9ics: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2',3'-DIDEOXYCYTIDINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DDCTP AND MNCL2 -
 9ict: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DGTP AND MNCL2
9ict: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DGTP AND MNCL2 -
 9icu: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DTTP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR)
9icu: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; SOAKED IN THE PRESENCE OF DTTP (1 MILLIMOLAR) AND MNCL2 (5 MILLIMOLAR) -
 9icv: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2'-DEOXYADENOSINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DATP AND ZNCL2
9icv: DNA POLYMERASE BETA (E.C.2.7.7.7)/DNA COMPLEX + 2'-DEOXYADENOSINE-5'-TRIPHOSPHATE, SOAKED IN THE PRESENCE OF DATP AND ZNCL2 -
 9icw: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; NATIVE STRUCTURE
9icw: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA; NATIVE STRUCTURE -
 9icx: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA (NON GAPPED DNA ONLY)
9icx: DNA POLYMERASE BETA (POL B) (E.C.2.7.7.7) COMPLEXED WITH SIX BASE PAIRS OF DNA (NON GAPPED DNA ONLY) -
 9icy: DNA POLYMERASE BETA (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA (NON GAPPED DNA ONLY)
9icy: DNA POLYMERASE BETA (E.C.2.7.7.7) COMPLEXED WITH SEVEN BASE PAIRS OF DNA (NON GAPPED DNA ONLY)
























































![2i9g: DNA Polymerase Beta with a Benzo[c]phenanthrene diol epoxide adducted guanine base](http://upload.wikimedia.org/wikipedia/commons/thumb/4/4c/PDB_2i9g_EBI.jpg/180px-PDB_2i9g_EBI.jpg)


















































































