Protein-coding gene in the species Homo sapiens
| APOC1 |
|---|
 |
| Available structures |
|---|
| PDB | Ortholog search: PDBe RCSB |
|---|
| List of PDB id codes |
|---|
1ALE, 1ALF, 1IOJ, 1OPP |
|
|
| Identifiers |
|---|
| Aliases | APOC1, Apo-CI, ApoC-I, apo-CIB, apoC-IB, apolipoprotein C1, Apolipoprotein C-I |
|---|
| External IDs | OMIM: 107710; MGI: 88053; HomoloGene: 136749; GeneCards: APOC1; OMA:APOC1 - orthologs |
|---|
| Gene location (Human) |
|---|
 | | Chr. | Chromosome 19 (human)[1] |
|---|
| | Band | 19q13.32 | Start | 44,914,247 bp[1] |
|---|
| End | 44,919,349 bp[1] |
|---|
|
| Gene location (Mouse) |
|---|
 | | Chr. | Chromosome 7 (mouse)[2] |
|---|
| | Band | 7 A3|7 9.94 cM | Start | 19,423,406 bp[2] |
|---|
| End | 19,426,585 bp[2] |
|---|
|
| RNA expression pattern |
|---|
| Bgee | | Human | Mouse (ortholog) |
|---|
| Top expressed in | - right lobe of liver
- nucleus accumbens
- putamen
- caudate nucleus
- right lung
- spleen
- substantia nigra
- hypothalamus
- lymph node
- upper lobe of left lung
|
| | Top expressed in | - left lobe of liver
- gallbladder
- lip
- brown adipose tissue
- yolk sac
- esophagus
- blastocyst
- subcutaneous adipose tissue
- white adipose tissue
- skin of back
|
| | More reference expression data |
|
|---|
| BioGPS | |
|---|
|
| Gene ontology |
|---|
| Molecular function | - phospholipase inhibitor activity
- lipase inhibitor activity
- phosphatidylcholine binding
- fatty acid binding
- phosphatidylcholine-sterol O-acyltransferase activator activity
| | Cellular component | - chylomicron
- very-low-density lipoprotein particle
- endoplasmic reticulum
- high-density lipoprotein particle
- extracellular region
| | Biological process | - lipid transport
- chylomicron remnant clearance
- negative regulation of fatty acid biosynthetic process
- lipid metabolism
- positive regulation of cholesterol esterification
- phospholipid efflux
- cholesterol metabolic process
- negative regulation of phosphatidylcholine catabolic process
- plasma lipoprotein particle remodeling
- negative regulation of very-low-density lipoprotein particle clearance
- regulation of cholesterol transport
- negative regulation of receptor-mediated endocytosis
- high-density lipoprotein particle remodeling
- cholesterol efflux
- negative regulation of cholesterol transport
- negative regulation of lipoprotein lipase activity
- very-low-density lipoprotein particle assembly
- negative regulation of lipid catabolic process
- triglyceride metabolic process
- negative regulation of lipid metabolic process
- positive regulation of catalytic activity
- very-low-density lipoprotein particle clearance
- lipoprotein metabolic process
| | Sources:Amigo / QuickGO |
|
| Orthologs |
|---|
| Species | Human | Mouse |
|---|
| Entrez | | |
|---|
| Ensembl | | |
|---|
| UniProt | | |
|---|
| RefSeq (mRNA) | |
|---|
NM_001645
NM_001321065
NM_001321066
NM_001379687 |
| |
|---|
| RefSeq (protein) | |
|---|
NP_001307994
NP_001307995
NP_001636
NP_001366616 |
| |
|---|
| Location (UCSC) | Chr 19: 44.91 – 44.92 Mb | Chr 7: 19.42 – 19.43 Mb |
|---|
| PubMed search | [3] | [4] |
|---|
|
| Wikidata |
| View/Edit Human | View/Edit Mouse |
|
| ApoC-I |
|---|
 structural studies of a baboon (papio sp.) plasma protein inhibitor of cholesteryl ester transferase. |
| Identifiers |
|---|
| Symbol | ApoC-I |
|---|
| Pfam | PF04691 |
|---|
| InterPro | IPR006781 |
|---|
| SCOP2 | 1ale / SCOPe / SUPFAM |
|---|
| Available protein structures: |
|---|
| Pfam | structures / ECOD |
|---|
| PDB | RCSB PDB; PDBe; PDBj |
|---|
| PDBsum | structure summary |
|---|
|
Apolipoprotein C-I is a protein component of lipoproteins that in humans is encoded by the APOC1 gene.[5][6]
Function
The protein encoded by this gene is a member of the apolipoprotein C family. This gene is expressed primarily in the liver, and it is activated when monocytes differentiate into macrophages. Alternatively spliced transcript variants have been found for this gene, but the biological validity of some variants has not been determined.[7]
Apolipoprotein C-I has a length of 57 amino acids normally found in plasma and responsible for the activation of esterified lecithin cholesterol with an important role in the exchange of esterified cholesterol between lipoproteins and in removal of cholesterol from tissues. Its main function is inhibition of cholesteryl ester transfer protein (CETP), probably by altering the electric charge of HDL molecules.
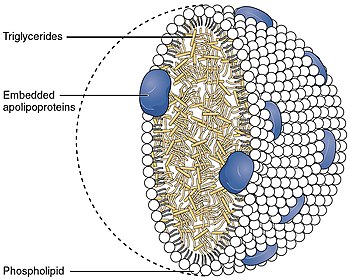
During fasting (like other apolipoprotein C), it is found primarily within HDL, while after a meal it is found on the surface of other lipoproteins. When proteins rich in triglycerides like chylomicrons and VLDL are broken down, this apoprotein is transferred again to HDL. It is one of the most positively charged proteins in the human body.
Pseudogene
A pseudogene of this gene is located 4 kb downstream from the apoC-I gene in the same orientation on chromosome 19, where both reside within an apolipoprotein gene cluster. This pseudogene, which was also reported to have been present in Denisovans and Neandertals, originated from two separate events. Following the divergence of New World monkeys from the human lineage, the apoC-I gene was duplicated. Old World monkeys and great apes other than humans have been shown to have two active genes. One of the duplicates encodes a basic protein designated apoC-IB that is orthologous to human apolipoprotein C-I. The other encodes an acidic protein, apoC-IA, that is orthologous to the virtual protein encoded by the pseudogene. The pseudogenization event occurred sometime between the divergence of bonobos and chimpanzees from the human lineage and the arrival of Denisovans and Neandertals. The pseudogene is due to a change in a single nucleotide in the codon for the penultimate amino acid, i.e. glutamine, in the signal sequence, resulting in a stop codon.[8][9][10]
Interactive pathway map
Click on genes, proteins and metabolites below to link to respective articles. [§ 1]
[[File:
|alt=Statin pathway edit]]
- ^ The interactive pathway map can be edited at WikiPathways: "Statin_Pathway_WP430".
References
- ^ a b c GRCh38: Ensembl release 89: ENSG00000130208 – Ensembl, May 2017
- ^ a b c GRCm38: Ensembl release 89: ENSMUSG00000040564 – Ensembl, May 2017
- ^ "Human PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ "Mouse PubMed Reference:". National Center for Biotechnology Information, U.S. National Library of Medicine.
- ^ Tata F, Henry I, Markham AF, Wallis SC, Weil D, Grzeschik KH, et al. (1985). "Isolation and characterisation of a cDNA clone for human apolipoprotein CI and assignment of the gene to chromosome 19". Human Genetics. 69 (4): 345–9. doi:10.1007/BF00291654. PMID 2985493. S2CID 32767041.
- ^ Smit M, van der Kooij-Meijs E, Frants RR, Havekes L, Klasen EC (January 1988). "Apolipoprotein gene cluster on chromosome 19. Definite localization of the APOC2 gene and the polymorphic Hpa I site associated with type III hyperlipoproteinemia". Human Genetics. 78 (1): 90–3. doi:10.1007/BF00291243. PMID 2892779. S2CID 22711986.
- ^ "Entrez Gene: APOC1 apolipoprotein C-I".
- ^ Puppione DL, Ryan CM, Bassilian S, Souda P, Xiao X, Ryder OA, Whitelegge JP (March 2010). "Detection of two distinct forms of apoC-I in great apes". Comparative Biochemistry and Physiology. Part D, Genomics & Proteomics. 5 (1): 73–9. doi:10.1016/j.cbd.2009.12.003. PMC 2830554. PMID 20209111.
- ^ Puppione D, Whitelegge JP (October 2013). "Proteogenomic Review of the Changes in Primate apoC-I during Evolution". Frontiers in Biology. 8 (5): 533–548. doi:10.1007/s11515-013-1278-7. PMC 5528196. PMID 28757862.
- ^ Puppione DL (September 2014). "Higher primates, but not New World monkeys, have a duplicate set of enhancers flanking their apoC-I genes". Comparative Biochemistry and Physiology. Part D, Genomics & Proteomics. 11: 45–8. doi:10.1016/j.cbd.2014.08.001. PMID 25160599.
Further reading
- Shulman RS, Herbert PN, Wehrly K, Fredrickson DS (January 1975). "Thf complete amino acid sequence of C-I (apoLp-Ser), an apolipoprotein from human very low density lipoproteins". The Journal of Biological Chemistry. 250 (1): 182–90. doi:10.1016/S0021-9258(19)41998-4. PMID 166984.
- Lauer SJ, Walker D, Elshourbagy NA, Reardon CA, Levy-Wilson B, Taylor JM (May 1988). "Two copies of the human apolipoprotein C-I gene are linked closely to the apolipoprotein E gene". The Journal of Biological Chemistry. 263 (15): 7277–86. doi:10.1016/S0021-9258(18)68638-7. PMID 2835369.
- Smit M, van der Kooij-Meijs E, Woudt LP, Havekes LM, Frants RR (May 1988). "Exact localization of the familial dysbetalipoproteinemia associated HpaI restriction site in the promoter region of the APOC1 gene". Biochemical and Biophysical Research Communications. 152 (3): 1282–8. doi:10.1016/S0006-291X(88)80424-8. PMID 2897845.
- Davison PJ, Norton P, Wallis SC, Gill L, Cook M, Williamson R, Humphries SE (May 1986). "There are two gene sequences for human apolipoprotein CI (apo CI) on chromosome 19, one of which is 4 kb from the gene for apo E". Biochemical and Biophysical Research Communications. 136 (3): 876–84. doi:10.1016/0006-291X(86)90414-6. PMID 3013172.
- Myklebost O, Rogne S (August 1986). "The gene for human apolipoprotein CI is located 4.3 kilobases away from the apolipoprotein E gene on chromosome 19". Human Genetics. 73 (4): 286–9. doi:10.1007/BF00279087. PMID 3017837. S2CID 11662593.
- Jackson RL, Sparrow JT, Baker HN, Morrisett JD, Taunton OD, Gotto AM (August 1974). "The primary structure of apolopoprotein-serine". The Journal of Biological Chemistry. 249 (16): 5308–13. doi:10.1016/S0021-9258(19)42365-X. PMID 4369340.
- Knott TJ, Robertson ME, Priestley LM, Urdea M, Wallis S, Scott J (May 1984). "Characterisation of mRNAs encoding the precursor for human apolipoprotein CI". Nucleic Acids Research. 12 (9): 3909–15. doi:10.1093/nar/12.9.3909. PMC 318798. PMID 6328444.
- Servillo L, Brewer HB, Osborne JC (February 1981). "Evaluation of the mixed interaction between apolipoproteins A-II and C-I equilibrium sedimentation". Biophysical Chemistry. 13 (1): 29–38. doi:10.1016/0301-4622(81)80022-1. PMID 6789904.
- Curry MD, McConathy WJ, Fesmire JD, Alaupovic P (April 1981). "Quantitative determination of apolipoproteins C-I and C-II in human plasma by separate electroimmunoassays". Clinical Chemistry. 27 (4): 543–8. doi:10.1093/clinchem/27.4.543. PMID 7471419.
- Rozek A, Buchko GW, Cushley RJ (June 1995). "Conformation of two peptides corresponding to human apolipoprotein C-I residues 7-24 and 35-53 in the presence of sodium dodecyl sulfate by CD and NMR spectroscopy". Biochemistry. 34 (22): 7401–8. doi:10.1021/bi00022a013. PMID 7779782.
- Maruyama K, Sugano S (January 1994). "Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides". Gene. 138 (1–2): 171–4. doi:10.1016/0378-1119(94)90802-8. PMID 8125298.
- Trask B, Fertitta A, Christensen M, Youngblom J, Bergmann A, Copeland A, et al. (January 1993). "Fluorescence in situ hybridization mapping of human chromosome 19: cytogenetic band location of 540 cosmids and 70 genes or DNA markers". Genomics. 15 (1): 133–45. doi:10.1006/geno.1993.1021. PMID 8432525.
- Kamino K, Yoshiiwa A, Nishiwaki Y, Nagano K, Yamamoto H, Kobayashi T, et al. (1996). "Genetic association study between senile dementia of Alzheimer's type and APOE/C1/C2 gene cluster". Gerontology. 42 (Suppl 1): 12–9. doi:10.1159/000213820. PMID 8804993.
- Rozek A, Buchko GW, Kanda P, Cushley RJ (September 1997). "Conformational studies of the N-terminal lipid-associating domain of human apolipoprotein C-I by CD and 1H NMR spectroscopy". Protein Science. 6 (9): 1858–68. doi:10.1002/pro.5560060906. PMC 2143781. PMID 9300485.
- Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S (October 1997). "Construction and characterization of a full length-enriched and a 5'-end-enriched cDNA library". Gene. 200 (1–2): 149–56. doi:10.1016/S0378-1119(97)00411-3. PMID 9373149.
- Halushka MK, Fan JB, Bentley K, Hsie L, Shen N, Weder A, et al. (July 1999). "Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis". Nature Genetics. 22 (3): 239–47. doi:10.1038/10297. PMID 10391210. S2CID 4636523.
- Freitas EM, Zhang WJ, Lalonde JP, Tay GK, Gaudieri S, Ashworth LK, et al. (1999). "Sequencing of 42kb of the APO E-C2 gene cluster reveals a new gene: PEREC1". DNA Sequence. 9 (2): 89–100. doi:10.3109/10425179809086433. PMID 10520737.
- Gautier T, Masson D, de Barros JP, Athias A, Gambert P, Aunis D, et al. (December 2000). "Human apolipoprotein C-I accounts for the ability of plasma high density lipoproteins to inhibit the cholesteryl ester transfer protein activity". The Journal of Biological Chemistry. 275 (48): 37504–9. doi:10.1074/jbc.M007210200. PMID 10978346.
- Hartley JL, Temple GF, Brasch MA (November 2000). "DNA cloning using in vitro site-specific recombination". Genome Research. 10 (11): 1788–95. doi:10.1101/gr.143000. PMC 310948. PMID 11076863.
External links
- Human APOC1 genome location and APOC1 gene details page in the UCSC Genome Browser.
- PDBe-KB provides an overview of all the structure information available in the PDB for Human Apolipoprotein C-I
PDB gallery
-
1ioj: HUMAN APOLIPOPROTEIN C-I, NMR, 18 STRUCTURES -
1opp: PEPTIDE OF HUMAN APOLIPOPROTEIN C-I RESIDUES 1-38, NMR, 28 STRUCTURES |
 | This article on a gene on human chromosome 19 is a stub. You can help Wikipedia by expanding it. |




 1ioj: HUMAN APOLIPOPROTEIN C-I, NMR, 18 STRUCTURES
1ioj: HUMAN APOLIPOPROTEIN C-I, NMR, 18 STRUCTURES 1opp: PEPTIDE OF HUMAN APOLIPOPROTEIN C-I RESIDUES 1-38, NMR, 28 STRUCTURES
1opp: PEPTIDE OF HUMAN APOLIPOPROTEIN C-I RESIDUES 1-38, NMR, 28 STRUCTURES